PromethION 2 Solo IT requirements
Requirements
PromethION 2 Solo IT requirements
FOR RESEARCH USE ONLY
Contents
This is a legacy document.
PromethION 2 Solo IT requirements
- 1. Overview
- 2. Using a GridION
- 3. Configuring a new computer
- 4. Storage requirements
- 5. Network access requirements
- 6. Other considerations
Frequently asked questions
Support
Change log
This is a legacy document.
Please refer to the PromethION 2 Solo – device and IT specifications document for guidance.
PromethION 2 Solo IT requirements
Overview
The PromethION™ 2 (P2) Solo is the smallest and most accessible product in the PromethION lineup. PromethION flow cells produce 6× more data than MinION™, making them ideal for larger genomes and high-plexity runs. The P2 Solo can run two flow cells simultaneously. For more information, click here.
The P2 Solo requires a computer with USB-C (USB 3.0+ speed). GridION™ Mk1 users can connect a P2 Solo to their sequencing device and use the GridION’s dedicated GPU and compute resources, or users can provide a compatible high-performance workstation.
Due to the large data volumes generated by the P2 Solo, selecting a suitably powerful computer is crucial. If you require an all-in-one solution with integrated compute and optimised performance, we recommend the P2 Integrated, which includes state-of-the-art computational hardware.
For more details on P2 Integrated, click here.
Using a GridION
The P2 Solo can use a GridION Mk1 as a plug-and-play compute resource, taking advantage of its integrated GPU for basecalling.
Out-of-the-box compatibility: The GridION’s operating system, SSD, and memory are fully optimised for sequencing with the P2 Solo.
Required GridION model: A GridION Mk1 with serial number GXB02xxx or higher is required to run all basecalling models with a PromethION Flow Cell.
For additional GridION-specific IT requirements, including network access and power requirements, click here.
Configuring a new computer
Selecting the right computer for P2 Solo is critical due to its higher data throughput. For optimal performance and installation simplicity, we recommend the P2 Integrated, which includes power compute hardware that is engineered for user experience.
The specifications below ensure your computer can efficiently acquire data and perform basecalling. Using an unsupported computer may lead to reduced performance, slower basecalling, or failed runs.
| Component | Specification |
|---|---|
| Operating System (OS) | Windows – 11 and 10 Linux – Ubuntu 22.04 and 24.04 |
| Peripheral | USB Type-C (USB 3.0 speeds or higher) |
| Memory | 64 GB+ |
| GPU | NVIDIA A100 NVIDIA 5090 RTX (Desktop card) RTX PRO 6000 Blackwell Workstation Edition |
| CPU | Intel i7+ (12-cores+) |
| Storage | 8 TB SSD + *Note if using multiple SSDs, they need to appear as a single drive to the OS (e.g. using RAID 0). |
Note: Compatibility and performance on newly released hardware, such as next-generation GPUs or processors, may be limited until testing and optimisation are complete.
Processor Compatibility
MinKNOW is only supported on modern Intel, AMD, and Apple Silicon processors.
Ensure your CPU supports AVX-2 (Intel Haswell, AMD Steamroller, or newer).
Intel-based Macs are not supported.
Other processors, such as Qualcomm Snapdragon or other ARM-based chips, are not supported.
USB Connectivity
The P2 Solo is designed to be connected via USB-C, and must be connected directly into the workstation computer's motherboard using the USB Type-C cable (~1 m) provided with the device.
Not supported:
PCIe to USB-C adapter cards (known to cause communication errors).
USB hubs or docking stations (may impact performance)
If your workstation lacks a USB-C port on the motherboard, a USB-C to USB-A cable can be provided. This USB-A port must support USB 3.0 speeds or higher.
Laptop suitability
NVIDIA offers graphics cards in both desktop and mobile (laptop) formats. While laptops provide portability, desktop cards typically offer better performance, power efficiency, and thermal management.
For compute-intensive tasks, desktops are generally more suitable, as laptops may struggle with sustained workloads.
Storage requirements
Nanopore sequencing data is stored in three file types:
FASTQ - A text-based format that stores DNA/RNA sequences along with quality scores.
BAM - A format for aligned reads, including modified base calls (e.g., methylation).
sequencing_summary.txt- Contains metadata for all basecalled reads from a sequencing run. This includes details such as read ID, sequence length, per-read Q-score, and read duration. The size of a sequence summary file will depend on the number of reads sequenced.
Optional:
- POD5 - The primary raw data format, replacing the legacy .fast5. It is more efficient in both storage and processing.
The table below provides estimated storage requirements based on different sequencing throughputs from a single flow cell. These values assume a run that saves POD5, FASTQ, and BAM files, with a read N50 of 23 kb.
| Flow cell output (Gbases) | POD5 storage (Gbytes) | FASTQ.gz storage (Gbytes) | Unaligned BAM with modifications (Gbytes) |
|---|---|---|---|
| 100 | 700 | 65 | 60 |
| 200 | 1,400 | 130 | 120 |
| 290 (TMO) | 2,030 | 188.5 | 174 |
Note: During sequencing, POD5 files are continuously generated. If basecalling is enabled, FASTQ and/or BAM files will be created, and POD5 writing may be disabled, however additional temporary storage will be required.
GridION data transfer
If using a P2 Solo with a GridION and requiring additional SSD storage, ensure you are using the correct USB/Ethernet connections for optimal performance.
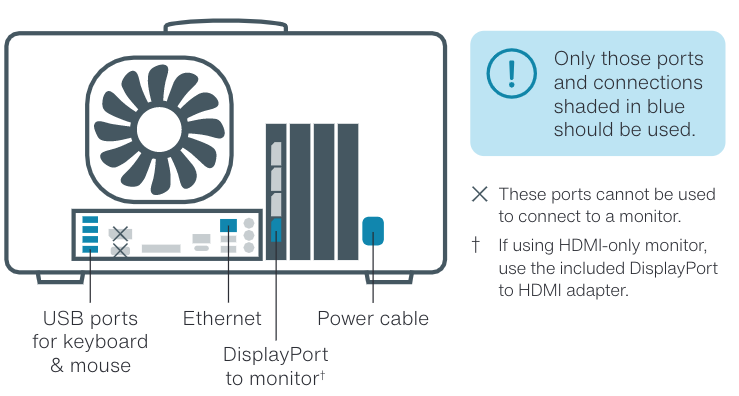
Use the blue USB Type-A ports on the rear of the GridION (see image above).
If using Ethernet, use a CAT5e or higher cable (1 Gbps minimum) and keep the cable as short as possible to reduce latency.
Do NOT use:
The USB port with a white rectangular centre
Any front-facing USB ports (if your GridION has them)
Network access requirements
Network access is required for correct functionality and system updates, all connections are outbound only over TCP ports 80 and 443. No inbound access is required.
Oxford Nanopore Technologies does not have remote access to your system.
| Access type | Purpose | Required domains |
|---|---|---|
| Telemetry | Enables MinKNOW to run and communicate telemetry | ping.oxfordnanoportal.com |
| Software & OS updates | Access to MinKNOW updates, OS packages, and GPU drivers | cdn.oxfordnanoportal.com .ubuntu.com .nvidia.com |
| EPI2ME™ Labs | Access for container-based analysis workflows | *.github.com hub.docker.com |
| Nanopore account login | Required to log in to your Nanopore account and access cloud services | id.nanoporetech.com *.okta.com |
If your institution uses a proxy or firewall, ensure outbound access to these domains is permitted to avoid issues with software functionality, updates, or user authentication.
Telemetry
MinKNOW and EPI2ME collect telemetry data during use as outlined in our Terms and Conditions. This helps monitor device performance, supports troubleshooting, and enables flow cell warranty replacement where applicable.
Privacy note: Some telemetry fields allow free-text entry, so avoid entering personally identifiable information. We do not collect sequence data.
EPI2ME analysis
The EPI2ME desktop application provides user-defined local or cloud-based analysis solutions:
Local analysis runs directly on the user’s computer, using available compute resources.
Cloud-based analysis is hosted on Amazon Web Services (AWS) and requires an internet connection.
Find out more about how EPI2ME can support your data analysis needs here.
Data upload formats: FASTQ, BAM, and other relevant workflow formats are uploaded through EPI2ME, which processes data via custom Nextflow pipelines and provides interactive HTML reports.
Other considerations
System behaviours such as scheduled updates, antivirus scans, or managed IT policies can interfere with sequencing runs.
Below are some key areas to review and discuss with your IT department to ensure a smooth and uninterrupted sequencing experience.
| Component | Minimum requirement |
|---|---|
| Electrical | Plug both the P2 Solo and the connected computer directly into wall power sockets. Do not use extension cords or power strips, as these can introduce electrical instability. |
| User account privilege level | Local Administrator privileges are required for installation and updates. They are not required for running sequencing experiments. |
| Internet connection | A stable internet connection is required at all times for software updates and telemetry. For offline use (e.g., fieldwork, expeditions), contact support@nanoporetech.com for guidance. |
| Antivirus settings | Antivirus software can consume significant system resources, potentially affecting sequencing performance. To avoid disruptions, we recommend disabling automatic scans and scheduling manual scans when the P2 Solo is not in use. |
| Endpoint Detection software | Endpoint detection software may interfere with system performance. We recommend disabling such software during sequencing. If your IT policies require third-party endpoint protection, please ensure that relevant codes are whitelisted in the software’s peripheral control settings. Users are responsible for validating that these settings do not affect P2 Solo performance. For more information please visit: How do I connect my P2 Solo? |
| BIOS/Chipset Settings & Updates | Ensure your BIOS firmware and chipset drivers are up to date. For EPI2ME analysis, ensure virtualisation is enabled. |
| OS update settings | Set OS updates to manual mode, as updates downloading during a sequencing run may cause issues. Updates requiring a restart will halt an active run Please coordinate with your IT department to ensure that Group Policies do not override local settings in a way that could cause unplanned updates or restarts. |
Frequently asked questions
Can I use a computer that differs from or is older than the specifications listed here?
Yes, but performance—especially basecalling speed—may vary depending on hardware. The choice of GPU has the greatest impact on performance.
Hardware compatibility:
Computers released before 2015 are unlikely to work.
Ensure your GPU supports CUDA 6.1+ (RTX 10XX series or newer).
Ensure your CPU supports AVX-2 (Intel Haswell, AMD Steamroller, or newer).
A USB-C port (USB 3.0 or higher) is required for device connection.
Newer hardware:
- Compatibility and performance on newly released hardware—such as next-generation GPUs or processors—may be limited until testing and optimisation are complete.
I bought a computer based on a previous version of this document, is it now obsolete?
This document is intended to guide users who are purchasing a new computer for use with the P2 Solo. As hardware availability changes over time, we update our recommendations to reflect current, readily available components. If your existing computer still meets the specifications listed in the question above, it remains compatible with our software.
I want to basecall my data on another computer / HPC, and want a true minimum specification for data acquisition only.
For data acquisition only (sequencing without any basecalling), the minimum specification - excluding the GPU - is sufficient.
Can I use Linux distributions other than Ubuntu 22.04/24.04 LTS?
We recommend using only Ubuntu 22.04 or 24.04 LTS. Other Linux distributions are not tested or validated and may not be compatible with our software or support processes.
Why isn’t there a recommended Mac specification?
While Apple Silicon is supported, sequencing performance is better on systems with NVIDIA GPUs, which also offer a more favourable price-to-performance ratio for basecalling. If using a computer powered by Apple Silicon, we only recommend use of fast basecalling, as larger models will take significant time to run and are not capable of keeping up with flow cell output.
Support
For more information and FAQs about the PromethION 2 Solo, refer to the P2 Solo self-service page.
Change log
| Date | Version | Changes made |
|---|---|---|
| 6th January 2026 | V15 | Document was moved to legacy. |
| 29 September 2025 | V14 | Information within each section was updated, which included the renaming of two sections and information within. Additional information was provided on recommended specifications for computers, network access requirements and required domains. Added new sections on 'Other considerations' for users to review with their IT department, and a section on 'FAQs'. Updated hardware recommendations to current Blackwell GPU equivalents. |
| 27th June 2025 | V13 | In the Table, under 'Configuring a new computer', '1 x USB-C port', the length of the USB Type-C cable was provided. |
| 2nd June 2025 | V12 | In 'Specifications' and 'Configuring a new computer', port 80 was added alongside port 443 for outbound IP addresses. |
| 8th May 2025 | V11 | The following note was added to the Table in section 3, 'Configuring a new computer' under GPU: Compatibility and performance on newly released hardware, such as next-generation GPUs or processors, may be limited until testing and optimisation are complete. |
| 31st July 2024 | V10 | - Renamed "Specifications - workstation/laptop" section to "Configuring a new computer." - Removed minimum specifications for Storage, Memory, GPU and CPU. - Updated recommended GPU to "NVIDIA GPU with at least 12GB of GPU memory. - Added note on GPU recommendations for real-time high accuracy basecalling. - Removed Apple silicon (M1, M2) GPU recommendation. - Added note on P2 Solo not appearing in MinKNOW due to antivirus/endpoint detection packages and how to resolve. - Merged "Example laptops" and "Example desktop" workstation lists to "Example computers" and updated items in list. - Renamed item "100-240 V 50/60 Hz AC power outlet" to "Electrical"; added recommendation to consult manufacturer electrical specifications - Added note not to use extension cord to connect P2 solo and computer. - Added note on operating temperatures of computer - In "File types", updated information about data generation for POD5, FASTQ and BAM files. |
| 24th April 2024 | V9 | - In "Specifications - workstation/laptop", updated operating system recommendations for Windows, macOS and Linux. - Updated CPU and GPU recommendations to include Apple silicon (M1 and M2) |
| 20th February 2024 | V8 | - In "Specifications - workstation/laptop", more information has been added about USB-C port requirements. - Minor corrections and clarifications throughout the document |
| 28th September 2023 | V7 | - A file size table has been added to "File types" - The required operating system for Mac has been updated in "Specifications - workstation/laptop" - Minor corrections and clarifications throughout the document |
| 4th August 2023 | V6 | A disclaimer has been added to "Specifications - workstation/laptop": "Do not use a USB-C to USB-A adapter to plug the device into your computer, as this can cause issues with connectivity." |
| 28th June 2023 | V5 | - In "Specifications - GridION" and "Specifications - workstation/laptop", the specs for telemetry feedback, EPI2ME analysis, and software updates have been changed. - In "Specifications - workstation/laptop", the minimum storage recommendations have been updated. - "Networking explanations" has been removed. - In "File types", information has been added about POD5 files and BAM files. The file size comparison table has been temporarily removed. - The "Included software" section has been removed. - A "Support" section has been added - Minor corrections and clarifications throughout the document. |
| 27th February 2023 | V4 | In "Specifications - workstation/laptop", "1x Type-C port" has been changed to "1x USB-C port" |
| 12th December 2022 | V3 | - Ubuntu 18.04 has been removed from the workstation/laptop specification table |
| 1st November 2022 | V2 | - A list of compatible workstations/laptops has been included after "Specifications - workstation/laptop" - In "Specifications - workstation/laptop", the requirements have been updated from 1 x USB TypeA/Type C port to 1 x USB Type-C port. |
| May 2022 | V1 | Initial publication |






